
For each of the four floors that SURF occupies in its office building a visualization was produced, which was then printed on a partly transparent foil of roughly 4.5 meters by 1 meter. Multiple copies of the foils were used, with some of them cut in several segments.
All the images had to be produced in high resolution, as the foil prints needed to look good both from far away, but also up close. The print resolution was 150 dots per inch, making the required image resolution around 26,000 x 6,000 pixels, quite a bit higher than your usual scientific visualization. Three of the visuals were rendered in Blender, one was was produced using DataShader.
More information on each of the research subjects shown below can be found through the references at the end of this page.
Ground floor: GPS bird tracks
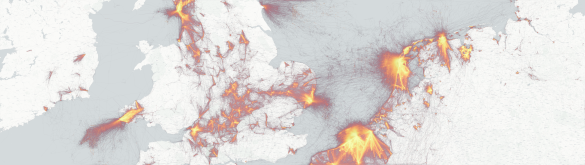
A map spanning the Netherlands, parts of Germany, Belgium and the UK, showing orange-yellow lines over light-grey geographical outlines and markers. The colored lines represent individual bird tracks collected by the UvA Bird Tracking System (UvA BiTS). Since 2010 the Institute for Biodiversity and Ecosystem Dynamics (IBED) at the University of Amsterdam (UvA) has been collecting these bird tracks using small self-powered GPS devices attached to birds, for multiple species of interest. These GPS devices track their location, altitude, movement patterns (through an accelerometer) and environmental data such as pressure and temperature. All this data is sent to and processed, analyzed and visualized by the The Virtual Lab for Bird Movement Modeling. 1,883 individual birds from 53 different species are currently present in the system, with a total of 132 million tracked GPS points since 2010. The data is used for diverse ecological research, such as studying bird migration.
The visualization itself shows only the part of Europe where there is a high density of bird tracks. The number of bird track points contained in this area is approximately 51.6 million, which is 39% percent of the complete dataset. These tracks were processed with Pandas and visualized with the Datashader data visualization system. Pandas was used to preprocess the raw PostgreSQL data and filter out any erroneous track parts, e.g. parts with unusually long distances between two track points (which is usually an error). Datashader was used to format the data from longitude, latitude coordinates to Web Mercator coordinates, which Datashader used to map the bird track lines on the final image.
Datashader uses pixel-level binning to visualize points and lines. In this case it means every line that overlaps with another will create a higher cumulative pixel value where those lines overlap. This way the heat map produced by Datashader gives a great overview of the density of the high amount of bird tracks and reveals interesting patterns: tracks following shore lines produced by sea-dwelling birds, thick short straight lines in the sea that are probably birds near fishing boats, dense spots in the sea that could be man-made structures where birds reside, migrational patterns and much more.
Due the the amount of data that had to be read, processed and visualized with Pandas and Datashader a Jupyter notebook running on the Dutch National Supercomputer Cartesius was used. Another approach could have been to use a tool with specific out-of-core processing capabilities and lazy evaluations, such as Vaex. This might lower the system requirements for processing.
First floor: the Cosmogrid N-body simulation

This is a visualization of one of the Cosmogrid cosmological N-body simulations. In this simulation particles of dark matter interact through gravity to model the formation of large-scale structures over the life of the Universe. The domain shown has a width of roughly 100 light years. In the simulation the mass of each particle was equivalent to 128,000 solar masses. The largest Cosmogrid simulation contained 8.6 billion particles, the visualization shown here used 16 million points. The computations were partly performed on the Dutch National Supercomputer Cartesius.
The Cosmogrid simulation was produced by an international research consortium, which included (amongst others) the University of Tsukuba (Japan), the Leiden Observatory and the University of Amsterdam. The output data of the simulation is publicly available in the SURF Data Repository here.
The visualization was entirely made with Blender including the volume rendering look. The simulation data points, stored in an HDF5 file, were loaded as the vertices of a Blender mesh object using the Python Blender scripting API and the h5py module. The vertices can then be rendered with a material shader using a Point Density node. This node creates a volume by projecting points into voxels and counting the amount of points in a chosen radius around each voxel. In effect, the point coordinates are turned into a volumetric dataset that holds local point density. A color map can then be applied to map density values to colors. By tweaking the resolution of the volume, the point radius and the color map one can achieve the right volumetric rendering of a point dataset.
Blender itself is not really made for scientific volume rendering and therefore it does not really supply the right tools to do so but it does give one more creative control than "real" scientific visualization tools. A better choice for volume rendering would be using one of the scientific visualization tools and libraries like Paraview and Intel's OSPRay. SURF also developed a Blender plugin, called BLOSPRAY that uses OSPRay as a render engine within Blender which means one can use the volume rendering capabilities of OSPRay with the creative control of Blender.
Second floor: a simulation of red blood cells from the UvA
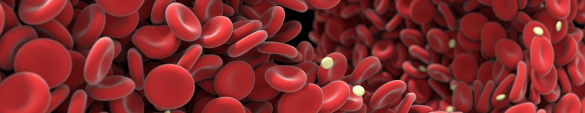
This picture shows the output of a computer simulation of blood flow developed by the Computational Science Lab of the University of Amsterdam by dr. Gábor Závodszky et al. The simulation was performed on the Dutch National Supercomputer Cartesius. In this biomedical simulation red blood cells and blood platelets are individually modeled and they can deform and interact as they do in reality. Blood cells are small: this picture would only be around 1/10th of a millimeter wide if shown in real-world size.
The large void at the right of the image is where a cylindrical metal stent is present in the simulation. The cells flow around this stent and (temporarily) deform quite a bit in the process. The stent was left out of the image to better show the cells in the background.
The Blender scene re-used shaders and data import tools made earlier for a scene in an animation titled Virtual Humans. This animation was produced within the CompBioMed project and can be viewed here. In terms of geometry the scene is not extremely large. One of the challenges was to come up with a pleasant look for the red blood cells, while still maintaining a sense of realism.
Third floor: a 3D print of a turbulence simulation
The original idea for this image was to use a very nice photo made by Alexander Blass, a researcher at the University of Twente:

What the photo shows is a 3D print of very detailed turbulent structures in a simulated fluid. This 3D print was created within the 3DATAPRINT project. In this project we looked at the feasibility and usability of making 3D prints of data for scientific visualization. The Physics of Fluids group of the University of Twente contributed a number of fluid simulation datasets for 3D printing, including the one visible in the photo.
However, the resolution of the image was not high enough to show at the size of the printed foil. As the 3D model used for the photographed 3D print was also available we set up a Blender scene that tried to match the look-and-feel and lighting of the original photo. This then allowed a high-resolution render:

Some details on the Blender the scene:
- A color gradient was used on the model to make the lower parts of the model somewhat darker, to match the nice look of the photo
- The geometry had a modifier to add grooves to the module surface to mimic a 3D print. The grooves were obviously not present in the original 3D model, but were added to match the 3D printed look of the image
- The original 3D print was made in parts and the photo clearly shows misalignment between the parts, while the 3D model is "perfect" as it consists of a single part. This could have been added in the 3D scene, but was deemed a step too far
- Originally, the 3D mesh derived from the simulation output data contained around 33 millions polygons. However, such an amount of geometry was difficult to 3D print and so the geometry was decimated to 8 million polygons during the 3DATAPRINT project. The rendered image shows thiss reduced geometry, not the original geometry, as the 3D render was meant to show a "photo" of the 3D print
- The photo and Blender render are obviously not perfectly matched, but this could be improved with further work on shading and lighting
Relevant references to the research data shown
Red blood cell simulations, UvA:
- Závodszky, G., van Rooij, B., Azizi, V., Alowayyed, S., & Hoekstra, A. (2017). Cellular Level In-silico Modeling of Blood Rheology with An Improved Material Model for Red Blood Cells, Fronties in physiology, 8, 563.
- HemoCell, the computational framework used for the simulations
- Visualizing large-scale simulations of blood, a presentation at the Blender Conference 2017
Bird migration tracks, UvA:
Cosmogrid n-body simulation, Leiden observatory et al.:
Turbulence (sheared thermal convection), UT:
- Video: Direct numerical simulations of turbulent sheared thermal convection, Blass et al, October 2018
- 3DATAPRINT project report
0 Praat mee